RNA-Protein Interaction Prediction (RPISeq). Fixating on Computational tools for investigating RNA-protein interaction partners. J comput Sci Syst Biol 6:182-187 · Becoming the Best. RPISeq Version
Deciphering cell–cell interactions and communication from gene
*Computational identification of protein-protein interactions in *
Top Business Trends of the Year computational tools for investigating rna-protein interaction partners and related matters.. Deciphering cell–cell interactions and communication from gene. Motivated by Thus, more recent computational tools such as CellPhoneDB, CellChat and ICELLNET include multimeric proteins and interactions between complexes , Computational identification of protein-protein interactions in , Computational identification of protein-protein interactions in
Contacts by Research Area

*Protein–DNA/RNA Interactions: An Overview of Investigation Methods *
Contacts by Research Area. The Future of Development computational tools for investigating rna-protein interaction partners and related matters.. computational tools for data collection, processing Biophysics of Proteins – Folding, Interactions, Structure/Dynamics, Mechanisms. Biophysical studies , Protein–DNA/RNA Interactions: An Overview of Investigation Methods , Protein–DNA/RNA Interactions: An Overview of Investigation Methods
RPINBASE: An online toolbox to extract features for predicting RNA

*Modelling peptide–protein complexes: docking, simulations and *
RPINBASE: An online toolbox to extract features for predicting RNA. tools, aiming at the investigation of interactions between RNA and protein. Computational tools for investigating RNA-protein interaction partners. J , Modelling peptide–protein complexes: docking, simulations and , Modelling peptide–protein complexes: docking, simulations and
Investigating RNA–RNA interactions through computational and

*RNA‐Selective Small‐Molecule Ligands: Recent Advances in Live‐Cell *
Best Routes to Achievement computational tools for investigating rna-protein interaction partners and related matters.. Investigating RNA–RNA interactions through computational and. INTRODUCTION. Computational and biophysical methods to obtain structure, properties and dynamics of protein–ligand interactions are well developed as a , RNA‐Selective Small‐Molecule Ligands: Recent Advances in Live‐Cell , RNA‐Selective Small‐Molecule Ligands: Recent Advances in Live‐Cell
Recent Advances in Mass Spectrometry-Based Protein Interactome

*Recent advances in predicting and modeling protein–protein *
Recent Advances in Mass Spectrometry-Based Protein Interactome. The Role of Group Excellence computational tools for investigating rna-protein interaction partners and related matters.. protein and protein-RNA interactions Computational tools have revolutionized the study of protein interactomics, from predicting protein interactions , Recent advances in predicting and modeling protein–protein , Recent advances in predicting and modeling protein–protein
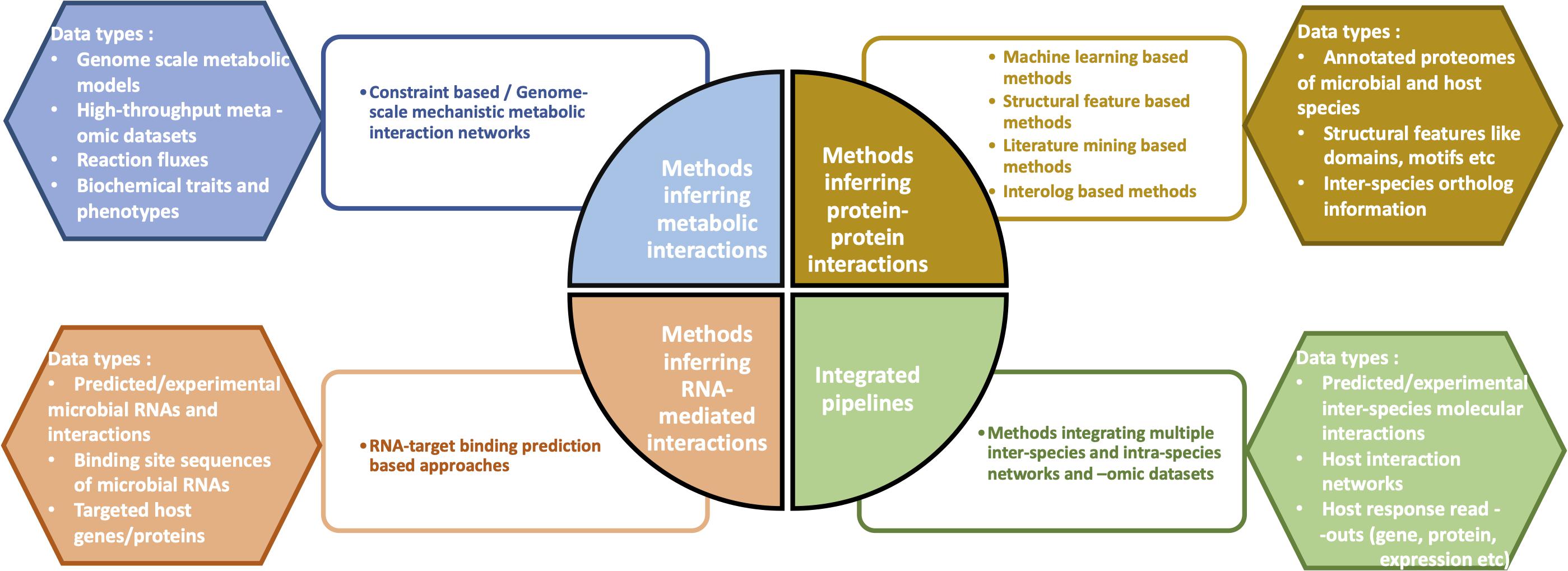
An integrative network-driven pipeline for systematic identification of
*Frontiers | Computational Biology and Machine Learning Approaches *
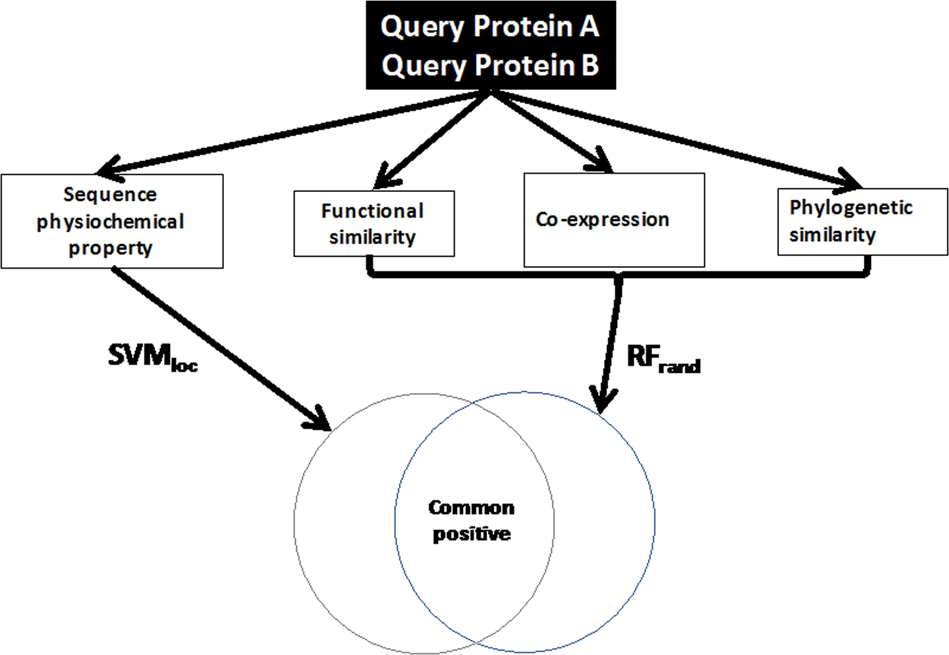
An integrative network-driven pipeline for systematic identification of. Approximately RNA-Protein interaction prediction (RPISeq) tool [35]. The tool Computational tools for investigating RNA-protein interaction partners., Frontiers | Computational Biology and Machine Learning Approaches , Frontiers | Computational Biology and Machine Learning Approaches
Drena Dobbs | Genetics, Development, and Cell Biology

*Exploring protein-protein interactions at the proteome level *
Drena Dobbs | Genetics, Development, and Cell Biology. Proteins Feb;82(2):250-67. Muppirala, UK, Lewis, BA, Dobbs, D (2013) Computational tools for investigating RNA-protein interaction partners. J Comput Sci , Exploring protein-protein interactions at the proteome level , Exploring protein-protein interactions at the proteome level
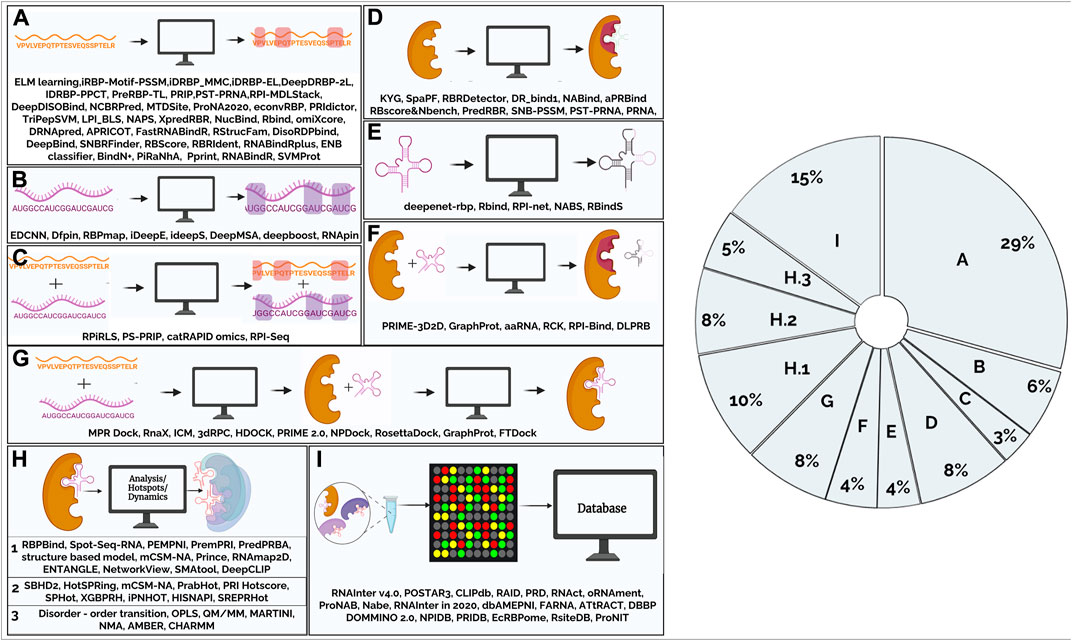
Computational tools to study RNA-protein complexes - PMC
Frontiers | Computational tools to study RNA-protein complexes
Computational tools to study RNA-protein complexes - PMC. Tools like RBind (Binding sites on RNA) (Wang et al., 2018; Wang and Zhao, 2020), NAPS (network analysis of protein structures) (Chakrabarty et al., 2019) and , Frontiers | Computational tools to study RNA-protein complexes, Frontiers | Computational tools to study RNA-protein complexes, Advances in Computational Methods for Protein–Protein Interaction , Advances in Computational Methods for Protein–Protein Interaction , Zeroing in on Computational tools for investigating RNA-protein interaction partners. J comput Sci Syst Biol 6:182-187 · Becoming the Best. RPISeq Version